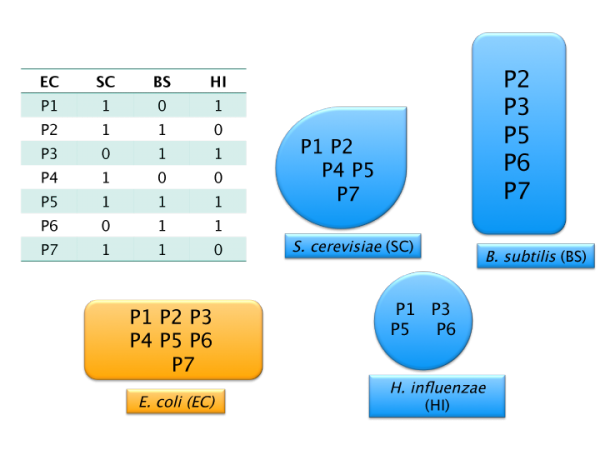
Phylogenetic profiling
Phylogenetic profiling uses homology information to describe a gene’s context in fully sequenced genomes (Pellegrini et al. 1999). It has been extended in numerous ways and successfully applied to deduce the function for a variety of genes (reviewed in Kensche et al. 2008).
A phylogenetic profile of a gene describes the pattern of presence/absence of its homologs across many genomes. This is informative of gene function because genes (and the corresponding proteins) that are gained and lost together tend to be involved in the same biological pathway - which would be incomplete without all its members in a given genome. In addition, the two genes with coordinated profiles may work in unrelated pathways that are both beneficial for fitness in a particular environment. Therefore, similarity of phylogenetic profiles suggests similarity of function.