Growth of sequence databases
GORBI uses machine learning to predict functions for millions of genes in 998 Bacteria and Archaea. You can search the database by the genome of interest, or by a particular Gene Ontology category, or by individual genes. Our work (Skunca et al. 2013) was motivated by the ever-widening gap between the large number of hypothetical proteins discovered by whole-genome sequencing, and the stagnating numbers of proteins whose function is characterized to some extent.
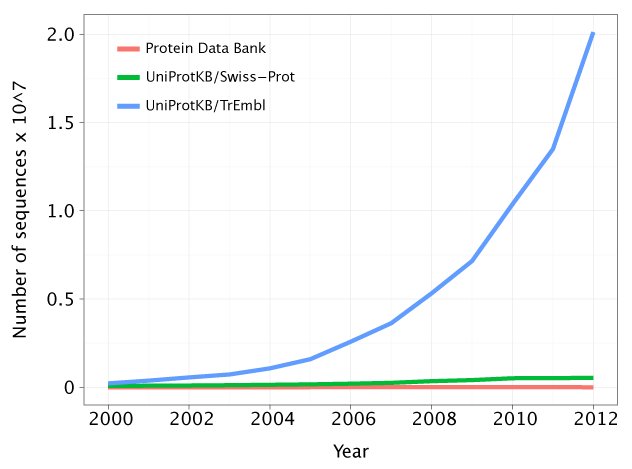
Data for PDB: http://www.wwpdb.org/stats.html
Data for UniProtKB/Swiss-Prot and UniProtKB/TrEmbl obtained form Claire O'Donovan via EBI database support
Because of the growing gap between the newly-sequenced and characterized sequences in the genome databases, computational methods in gene functional annotation are indispensable. Moreover, given the drop in the cost of the genome sequencing techniques, this gap is only destined to grow.
It is therefore useful to have a set of computational predictions on which one can focus the effort. It is our hope that the results of computational functional annotation presented in GORBI will be used in directing experiments toward smaller subsets of targets, thereby increasing the cost/benefit ratio of every experiment performed.