Experimental validation of the model's accuracy estimates
To validate how realistic are the confidence estimates reported by our machine learning model, we chose annotations for 38 genes in Escherichia coli K-12 at a threshold of 60% expected Precision.
We focused on three GO terms that were straightforward to investigate experimentally using readily available antibiotics:
- response to DNA damage stimulus,
- translation,
- peptidoglycan-based cell wall biogenesis.
The 38 E. coli strains, each with the deletion of one selected gene, were grown in the presence of antibiotics that target the above processes:
- nalidixic acid (causes severe DNA damage),
- kasugamycine (inhibitor of translation initiation),
- and ampicillin (inhibitor of cell wall synthesis).
We considered a prediction confirmed only if: (1) the survival of a mutant was <25% of the wild-type, when grown with the antibiotic inhibiting the process GORBI has predicted, and (2) the survival of the mutant was >50% of the wild-type when grown on the other two antibiotics, here used as negative controls.
For example, we predicted “DNA damage response” for the E. coli yfgI gene: when grown with DNA-damaging nalidixic acid, the yfgI mutant had 7% survival of the wild type, but when grown on kasugamycine or ampicilin, the survival was much higher: 98% and 74% of the wild type, respectively. We therefore consider this prediction confirmed: the yfgI mutant is sensitive to a DNA-damaging agent, while exhibiting wild type-like resistance to other stresses.
With these criteria, 25 out of 38 genes had confirmed predictions, equivalent to an experimental Precision of 66% (95% C.I. 51%-81%). This agrees well with the expected Precision of ≥60%, show that our estimates of accuracy are realistic.
In fact, 14 of the 38 tested genes have Precision ≥85%. For these genes, the experiments have shown 11/14 (79%) to be correct, approximately matching the expected precision of 85%.
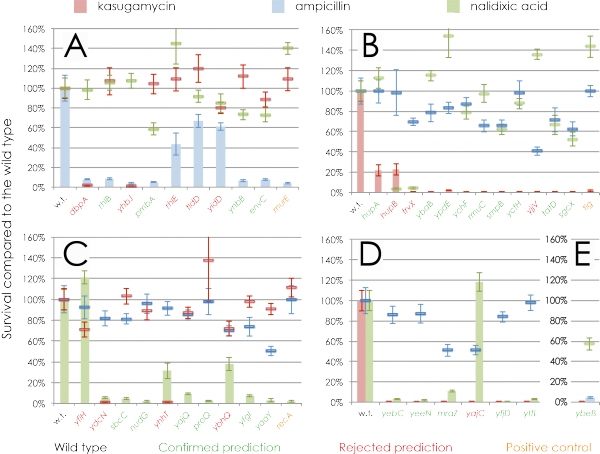
Experimental validation of predictions. A) Genes predicted to be annotated with "peptidoglycan-based cell wall biogenesis," B) genes predicted to be annotated with "translation," C) genes predicted to be annotated with "response to DNA damage stimulus," D) genes predicted to be annotated with both "translation" and "response to DNA damage stimulus," and E) a gene predicted to be annotated with both "translation" and " peptidoglycan-based cell wall biogenesis." The x-axis denotes the Escherichia coli knockout mutant. The y-axis represents the percentage of survival of the mutant strain normalized to the wild type. Coloured bars represent the survival when the antibiotic disrupts the biological process we predict for the genes; here, the correctly annotated mutants are expected to survive less than the wild type (w. t.). Coloured lines represent the survival when we predict no effect of the antibiotic on the survival rate; here, deletion mutants were expected not to differ from the wild type. Error bars show the variation in the results among the four replicates.