Combining orthologs and paralogs to accurately predict gene function
The GORBI database hosts the results of a model for computational assignment of gene function that builds on recent research: while orthologs are significantly more similar in function than paralogs, the overall difference is surprisingly modest (Altenhoff et al, PLoS Comp Biol 2012) and many paralogs have very conserved function (Nehrt et al, Plos Comp Biol 2011).
The result is a machine learning framework for orthology and paralogy-aware phylogenetic profiling that provides a large number of computational annotations with high accuracy in train/test evaluations.
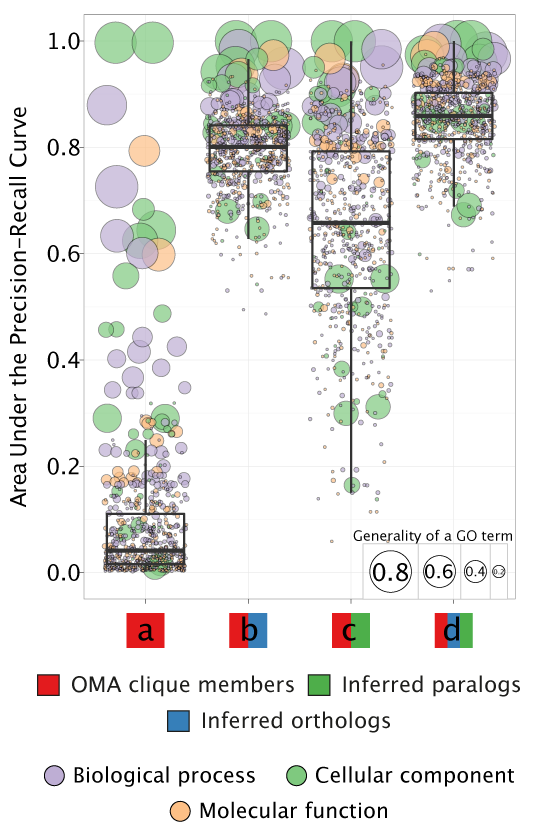
In one OMA clique, inferred orthologous relations connect each protein to every other protein. Consequently, OMA cliques leave out many of the existing orthologous relations. This makes the phylogenetic profiles of OMA cliques incomplete, leading to poor performance in our classification model: it can successfully annotate using only the most general GO terms (model a).
If we compensate by adding orthologs that were left out when constructing the original OMA cliques, the model improves: the mean AUPRC is 0.8 (model b). This includes all one-to-one, one-to-many, many-to-one, and many-to-many orthology relationships.
We also tested whether adding paralogs to phylogenetic profiles of OMA cliques improves the mean AUPRC: it does, showing that the paralogs are also a valuable source of information about gene function (model c). Still, the mean AUPRC is 0.65—lower than if we construct phylogenetic profiles with orthology relationships instead of the paralogs.
It is, however, the combined pattern of the orthologs and paralogs together that provides us with the best model for functional annotation (model d): the mean AUPRC increases to 0.85.
Therefore, all the results available in GORBI are predictions of the model d.