How to use: a tutorial
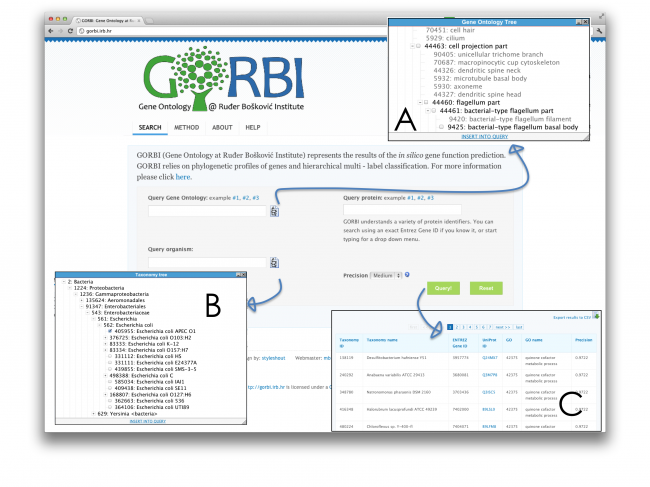
Predictions can be browsed in three ways, and any of their combination:
1) query using a Gene Ontology (GO) accession number (e.g. 6418 for "tRNA aminoacylation for protein translation"); alternatively, one can browse the "Gene Ontology tree" (insert A) to find interesting GO terms.
2) query using an organism's NCBI taxonomy ID (e.g. 288681 for Bacillus cereus E33L); alternatively, one can browse the "Taxonomy tree" (insert B) to find the NCBI taxonomy IDs of interesting organisms.
3) query using protein identifiers: NCBI Gene ID, UniProtKB protein ID, RefSeq ID, UniRef ID, UniParc, or EMBL ID.
The results (insert C) list NCBI taxonomy ID and the name of the organism, Entrez Gene ID, UniProt ID that links to the corresponding protein's page in the UniProt Knowledge Base, GO accession number, the name and the expected precision for the prediction.